Welcome to lcr-modules’s Documentation!¶
Getting Started With lcr-modules¶
- Users: Check out this Getting Started guide and the Demo Project.
- Contributors: Check out this Getting Started guide and the lcr-modules repository.
Installing oncopipe¶
You can install the latest version of oncopipe released on the Python Package Index using pip.
pip install oncopipe
Alternatively, you can install the development version of oncopipe from the lcr-modules repository. This way, your installation will automatically update when you pull from GitHub.
git clone https://github.com/LCR-BCCRC/lcr-modules.git
pip install -e lcr-modules/oncopipe
Motivation¶
This project aims to become a collection of standard analytical modules for genomic and transcriptomic data. Too often do we copy-paste from each other’s pipelines, which has several pitfalls:
* Too much time spent on routine analyses * Increased risk for hidden logical bugs
* Duplicated effort within and between labs * No consistently used pipelining tool
* Inefficient dissemination of best practices * Steep learning curve for new members
Fortunately, all of these problems can be solved with standardized analytical modules, and the benefits are many:
* Projects can ramp up faster * Consistent intermediate/output files
* Streamline efforts between labs * More reproducible analyses
* Define analytical best practices * Easier-to-write methods
* Consolidate collective expertise * Automated logging and “paper trail”
* Simplify member onboarding * Easier peer review of code
* And happier bioinformaticians!
What Are Modules?¶
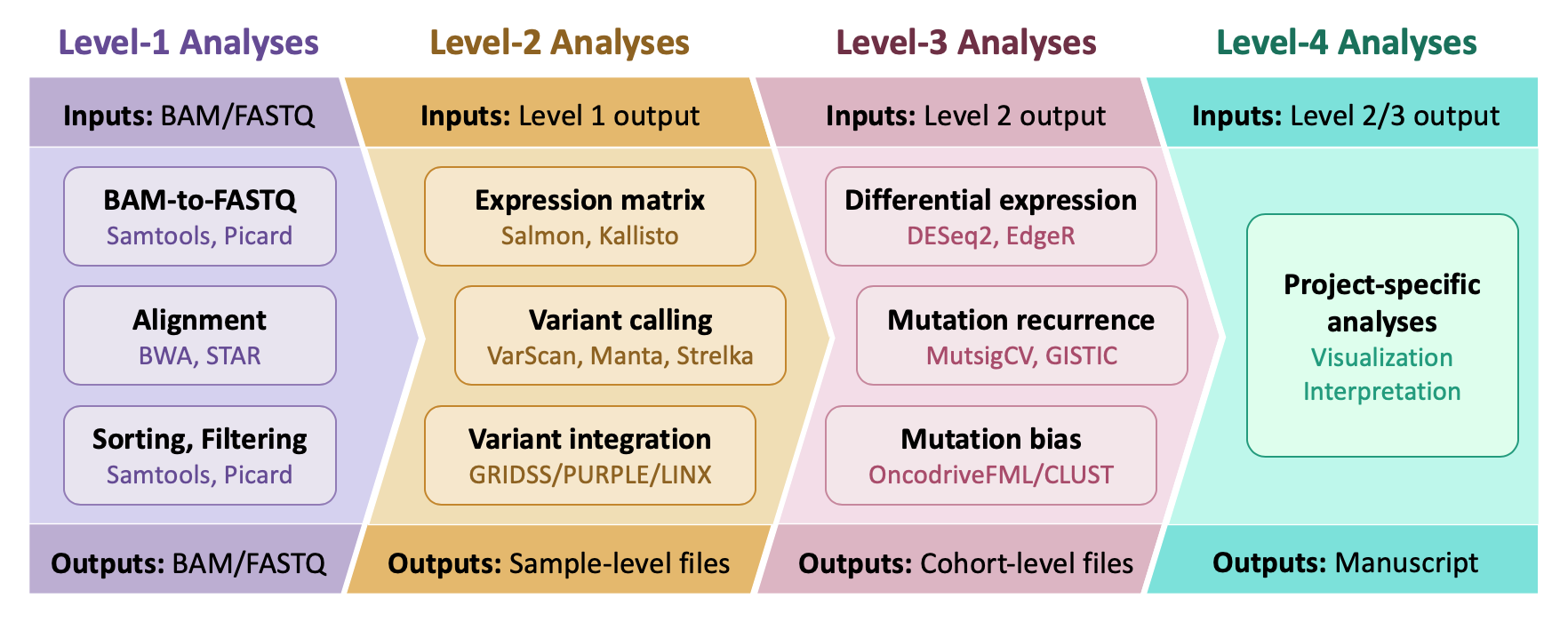
Each module accomplishes a specific analysis, generally centered around a specific tool (e.g. Strelka2, Manta, MutSigCV). Analyses—and by extension, modules—can be organized into different levels. The figure below contains for examples for each level.
- Level-1 Analyses: They process raw sequencing data, generally producing BAM/FASTQ files.
- Level-2 Analyses: They perform sample-level analyses on level-1 output, such as variant calling and gene expression quantification.
- Level-3 Analyses: They aggregate sample-specific level-2 output and perform cohort-wide analyses, such as the identification of sifgnificantly mutated genes.
- Level-4 Analyses: They are project-specific and are meant to ask specific questions of the data. These are the analyses you ideally want to spend your time on.